Autophagie and Cancer
Institute of PathologyUniversity of Bern
Murtenstrasse 31
CH-3008 Bern, Switzerland - Bern
Site web - mario.tschan@pathology.unibe.ch - 41316328780
Principal investigator
Mario P. Tschan

Research themes
The members of my research team share the interest to identify the regulation and function of autophagy in acute myeloid leukemias, breast, esophageal and lung cancer pathology. In close collaboration with clinical pathologists, we decipher the molecular mechanisms of the aberrant autophagy recycling pathways found in these tumor types. Moreover, we investigate the role of autophagy in resistance mechanisms towards chemo- or targeted therapies.
– Molecular analysis of autophagy pathways in acute myeloid leukemia (AML) differentiation therapy
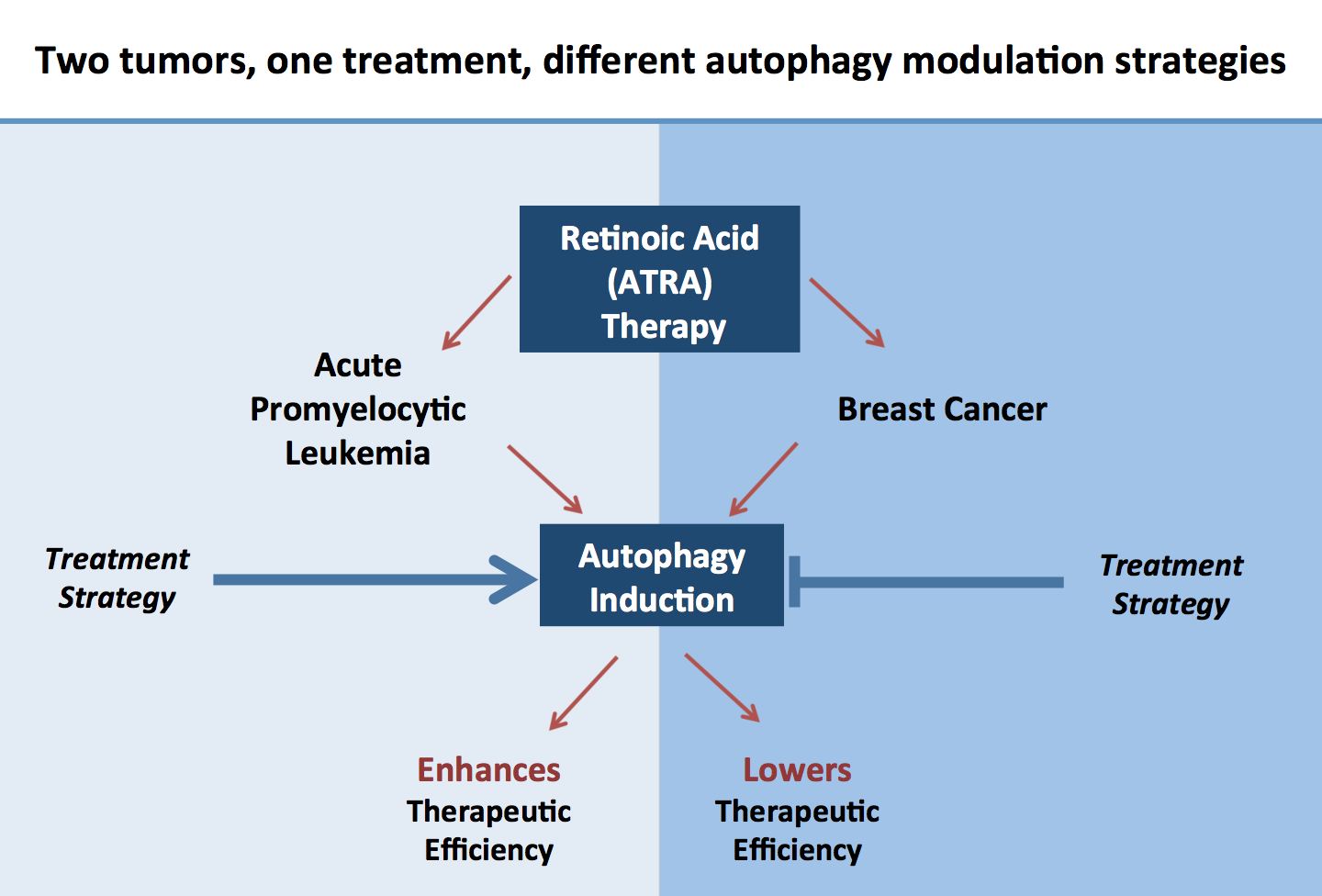
Retinoids are naturally occurring vitamin A derivatives, which exert their functions via activation of nuclear retinoid acid receptor mediated gene expression. All-trans retinoic acid (ATRA) is successfully used to treat acute promyelocytic leukemia (APL) by inducing neutrophil differentiation of leukemic blast cells. We and others reported on significantly increased macroautophagy activity during ATRA treatment as well as impaired granulocytic differentiation upon pharmacological or genetic inhibition of autophagy in APL cells. Accordingly, we found significantly decreased expression of key autophagy genes in primary AML patients as compared granulocytes from healthy donors. Importantly, pharmacological activation of autophagy in combination with ATRA treatment significantly boosted APL differentiation. Generally, our data provide strong evidence for a particular, Beclin-1 and ATG16L1-independent non-canonical subtype of autophagy operative during neutrophil differentiation of APL cells.
Another interest in my laboratory is to understand how Death Associated Kinase 2 (DAPK2) contributes to therapy-induced autophagy in APL. Our recent findings indicate that two different pathways are operative upon ATRA or arsenic trioxide (ATO) treatment. We found that DAPK2 is essential for ATRA- but not ATO-mediated autophagy while it is necessary for ATO-induced cell death responses.
Lastly, in a project lead by Magali Humbert we investigate the involvement of chaperone-mediated autophagy (CMA) and its interplay with non-canonical macroautophagy during APL differentiation therapy. This project also includes studies on how the leukemic niche affects autophagy in AML cells.
– Retinoic acid therapy and autophagy in breast cancer
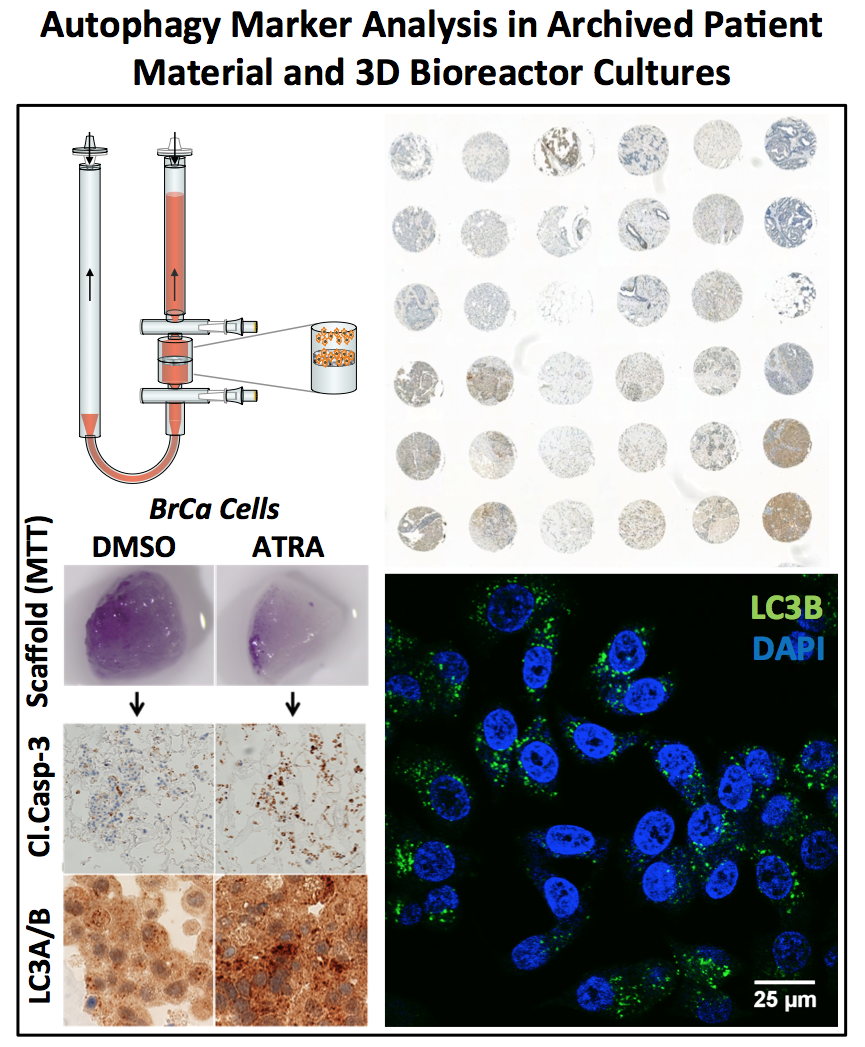
Clinical studies using ATRA for breast cancer therapy were rather disappointing and a better understanding of the ATRA-induced toxicity and differentiation of breast cancer cells is needed. We found that ATRA induces a significant increase in autophagic flux in breast cancer cells expressing the retinoic receptor alpha. Blocking autophagy in combination with ATRA treatment resulted in a marked increase of apoptosis in breast cancer cells. Importantly, autophagy can support the acquisition of epithelial traits of normal and cancer cells. Our current studies lead by Anna Schläfli aim at analyzing the function of autophagy in epithelial differentiation of breast cancer cells to improve retinoid therapy.
Descriptive figure
Publications
1. Jin J, Britschgi A, Schläfli AM, Humbert M, Shan-Krauer D, Batliner J, Federzoni EA, Ernst M, Torbett BE, Yousefi S, Simon HU, Tschan MP. Low Autophagy (ATG) Gene Expression Is Associated with an Immature AML Blast Cell Phenotype and Can Be Restored during AML Differentiation Therapy. Oxid Med Cell Longev, Volume 2018, Article ID 1482795.
2. Schläfli AM, Isakson P, Garattini E, Simonsen A, Tschan MP.The autophagy scaffold protein ALFY is critical for the granulocytic differentiation of AML cells. Sci Rep. 2017 Oct 11;7(1):12980.
3. Humbert M, Federzoni EA, Tschan MP. Distinct TP73-DAPK2-ATG5 pathway involvement in ATO-mediated cell death versus ATRA-mediated autophagy responses in APL. J Leukoc Biol. 2017 Dec;102(6):1357-1370.
4. Niklaus M, Adams O, Berezowska S, Zlobec I, Graber F, Slotta-Huspenina J, Nitsche U, Rosenberg R, Tschan MP, Langer R. Expression analysis of LC3B and p62 indicates intact activated autophagy is associated with an unfavorable prognosis in colon cancer. Oncotarget. 2017 May 2;8(33):54604-54615.
5. Rothschild SI, Gautschi O, Batliner J, Gugger M, Fey MF, Tschan MP. MicroRNA-106a targets autophagy and enhances sensitivity of lung cancer cells to Src inhibitors. Lung Cancer. 2017 May;107:73-83.
6. Berezowska S, Galván JA. Immunohistochemical Detection of the Autophagy Markers LC3 and p62/SQSTM1 in Formalin-Fixed and Paraffin-Embedded Tissue.
Methods Mol Biol. 2017;1560:189-194.
7. Adams O, Dislich B, Berezowska S, Schläfli AM, Seiler CA, Kröll D, Tschan MP, Langer R. Prognostic relevance of autophagy markers LC3B and p62 in esophageal adenocarcinomas. Oncotarget. 2016 Jun 28;7(26):39241-39255.
8. Schläfli AM, Adams O, Galván JA, Gugger M, Savic S, Bubendorf L, Schmid RA, Becker KF, Tschan MP, Langer R, Berezowska S. Prognostic value of the autophagy markers LC3 and p62/SQSTM1 in early-stage non-small cell lung cancer. Oncotarget. 2016 Jun 28;7(26):39544-39555.
9. Schläfli AM, Berezowska S, Adams O, Langer R, Tschan MP. Reliable LC3 and p62 autophagy marker detection in formalin fixed paraffin embedded human tissue by immunohistochemistry. Eur J Histochem. 2015 May 5;59(2):2481.
10. Brigger D, Schläfli AM, Garattini E, Tschan MP. Activation of RARα induces autophagy in SKBR3 breast cancer cells and depletion of key autophagy genes enhances ATRA toxicity. Cell Death Dis. 2015 Aug 27;6:e1861.
Composition de l'équipe
Magali Humbert, postdoc, magali.humbert@pathology.unibe.ch
Félice Janser, PhD student, ariane.janser@pathology.unibe.ch
Deborah Krauer, laboratory technician, deborah.shan@pathology.unibe.ch
Sophie Milesi, Master student, sophie.milesi@pathology.unibe.ch
Severin Mosimann, Master student, severin.mosimann@pathology.unibe.ch
Nicolas Niklaus, PhD student, nicolas.niklaus@pathology.unibe.ch
Sarah Parejo, Laboratory technician, sarah.parejo@pathology.unibe.ch
Sreoshee Rafiq, PhD student, sreoshee.rafiq@pathology.unibe.ch
Kristina Seiler, MD-PhD student, kristina.seiler@pathology.unibe.ch
Anna Schläfli Bill, postdoc, anna.schlaefli@pathology.unibe.ch
Igor Tokarchuk, MD-PhD student, igor.tokarchuk@pathology.unibe.ch
Mario P. Tschan, Associate Professor, mario.tschan@pathology.unibe.ch
Collaborations avec des cliniciens
Rupert Langer, MD, Associate Professor, rupert.langer@pathology.unibe.ch
Sabina Berezowska, MD. Assistant Professor, sabina.berezowska@pathology.unibe.ch