M2iSH (Microbes, Intestin, Inflammation and Susceptibility of the Host)
Inserm UMR 1071, INRAE USC 2018, Université Clermont Auvergne.Adresse : 28 Place Henri Dunant, CBRV, 63000 Clermont–Ferrand, France. - Clermont–Ferrand
Site web - hang.nguyen@uca.fr - 0473177997
Principal investigator
Dr. Hang NGUYEN

Research themes
Axis 1: Infectious etiology of Crohn’s disease and colorectal cancer
The UMR 1071 research unit has been working on the interaction between the host and pathogenic bacteria, in particular the pathogenic Escherichia coli involved in inflammatory bowel diseases, in particular Crohn’s disease (CD), and colorectal cancer (CRC).
The UMR 1071 was pioneer in discovering the abnormal colonization of the ileal mucosa from CD patients with adherent-invasive E. coli (AIEC) strains. Our hypothesis is that the development of CD could be linked to a genetic susceptibility of patients to be colonized by AIEC, which are able to adhere to and invade intestinal epithelial cells, to survive and replicate in macrophages, and to induce a pro-inflammatory response. Since then, the role of AIEC bacteria in CD has been confirmed by many other research groups around the world.
As a strong link between chronic inflammation and cancer has been established, the UMR 1071 has also developed a research axis aiming at investigating the infectious etiology of CRC, in particular the role of pathogenic E. coli in this disease. We have shown that E. coli strains producing colibactin (“colibactin-producing E. coli” or CoPEC), a DNA damage-inducing toxin, play a role in tumor progression.
Within the UMR 1071 research unit, we are developing three main research objectives, which aim to: (i) explore the virulence of E. coli strains involved in the etiology of chronic intestinal pathologies, (ii) study the predisposition of the host to E. coli infection in CD and CRC, and (iii) develop new biomarkers and specific therapies based on modulation of intestinal microbiota.
Axis 2: Autophagy and the E. coli strains associated to CD and CRC (AIEC and CoPEC strains)
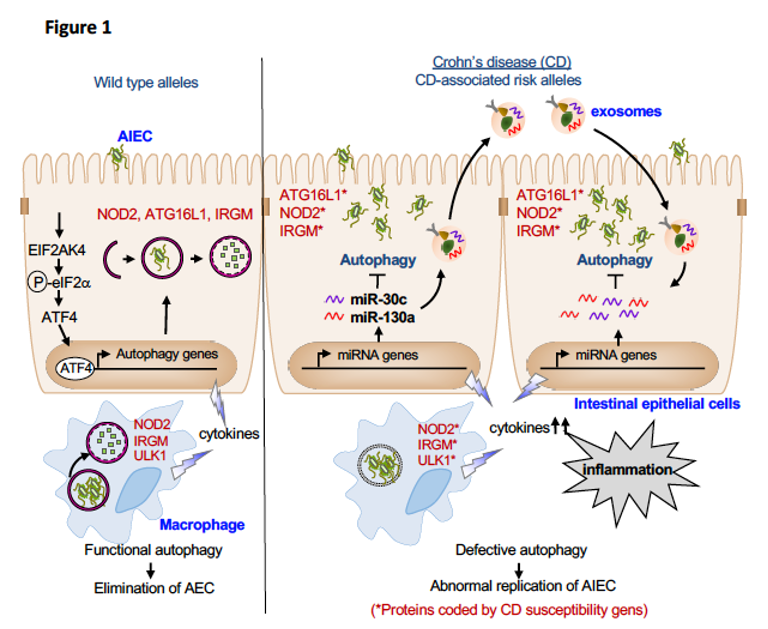
Dysfunctional autophagy is recognized as a contributing factor in many chronic inflammatory diseases, including inflammatory bowel diseases. The works of the UMR 1071 research unit have shown that autophagy plays a central role in the control of AIEC intracellular replication. We have shown that the CD-associated polymorphisms in the autophagy-related genes lead to defective autophagy, thus promoting the intracellular persistence of AIEC and an exacerbated pro-inflammatory response.
We have also shown that activation of the metabolic stress response pathway EIF2AK4-EIF2A-ATF4 upon AIEC infection serves as a host defense mechanism to induce a functional autophagy, independently of mTOR, to control AIEC intracellular replication and to inhibit AIEC-induced inflammation. Given that most of the current strategies based on modulating autophagy by targeting mTOR can have undesirable effects, targeting the EIF2AK4-EIF2A-ATF4 pathway could be a promising strategy to inhibit specifically AIEC colonization and inflammation.
While host cells induce a functional autophagy to control AIEC replication, AIEC can subvert autophagy by up-regulating the levels of miR-30c and miR-130a in intestinal epithelial cells, thus inhibiting the levels of ATG5 and ATG16L1. AIEC also inhibit autophagy by impairing host SUMOylation, a eukaryotic-reversible post-translational modification. The impaired autophagy induced by AIEC via the modulation of host miRNAs or SUMOylation consequently leads to increases in AIEC intracellular replication and inflammation.
Recently, we showed that upon AIEC infection, intestinal epithelial cells and macrophages secrete an increased amount of exosomes, which are extracellular vesicles of 30 to 100 nm playing a role in cell-to-cell communication. Furthermore, the exosomes secreted by AIEC-infected cells can transfer miR-30c and miR-130a from cell-to-cell, thus inhibiting autophagy in exosome-receiving cells and consequently impairing AIEC clearance.
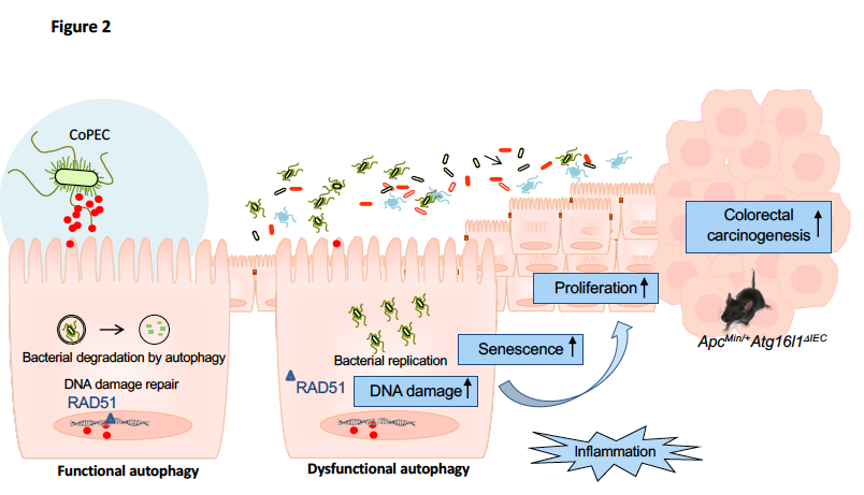
The link between autophagy and the bacteria implicated in the etiology of CRC remains largely unknown. Our recent study focuses on the role of autophagy in host defense to CoPEC infection. We showed that the presence of CoPEC strains in the colonic mucosa from patients with sporadic CRC is correlated with high mRNA expression levels of autophagy-associated genes. Autophagy is activated in host intestinal epithelial cells upon CoPEC infection in vitro and in vivo, and this is necessary to inhibit the pro-tumoral and pro-inflammatory effects of the bacteria, thus suppressing the colorectal carcinogenesis induced by the latter in a mouse model of CRC, the ApcMin/+ mice.
Together, our studies will provide a better understanding of the role of autophagy in host defense in the control of E. coli strains associated with CD and CRC. In the longer term, these works could contribute to the development of novel therapeutic strategies for patients with abnormal colonization of AIEC or CoPEC based on modulation of autophagy.
Descriptive figure
Publications
1. Lucas C, Salesse L, Hoang MHT, Bonnet M, Sauvanet P, Larabi A, Godfraind C, Gagnière J, Pezet D, Rosenstiel P, Barnich N, Bonnet R, Dalmasso G, Nguyen HTT. Autophagy of intestinal epithelial cells inhibits colorectal carcinogenesis induced by colibactin-producing Escherichia coli in ApcMin/+ mice. Gastroenterology. 2020, 158(5):1373-1388. PMID: 31917256.
2. Larabi A, Dalmasso G, Delmas J, Barnich N, Nguyen HTT. Exosomes transfer miRNAs from cell-to-cell to inhibit autophagy during infection with Crohn’s disease-associated adherent-invasive E. coli. Gut Microbes. 2020, 1; 11(6):1677-1694. PMID: 32583714.
3. Buisson A, Douadi C, Ouchchane L, Goutte M, Hugot JP, Dubois A, Minet-Quinard R, Bouvier D, Bommelaer G, Vazeille E, Barnich N. Macrophages inability to mediate adherent-invasive E. coli replication is linked to autophagy in Crohn’s disease patients. Cells. 2019, 5; 8(11):1394. PMID: 31694333
4. Larabi A, Barnich N, Nguyen HTT. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy. 2019; 9:1-14. PMID: 31286804.
5. Dalmasso G Nguyen HTT, Faïs T, Massier S, Barnich N, Delmas J, Bonnet R. Crohn’s disease-associated adherent-invasive Escherichia coli manipulate host autophagy by impairing SUMOylation. Cells. 2019; 9; 8(1). PMID: 30634511.
6. Bretin A, Lucas C, Larabi A, Dalmasso G, Billard E, Barnich N, Bonnet R, Nguyen H*. AIEC infection triggers modification of gut microbiota composition in genetically predisposed mice, contributing to intestinal inflammation. Scientific Reports. 2018; 8:12301. PMID: 30120269.
7. Carrière J, Bretin A, Darfeuille-Michaud A, Barnich N, Nguyen HT. Exosomes released from cells infected with Crohn’s disease-associated adherent-invasive Escherichia coli activate host innate immune responses and enhance bacterial intracellular replication. Inflammatory Bowel Disease. 2016, 22(3):516-28. IF2014: 4.46.
8. Bretin A, Carrière J, Dalmasso G, Bergougnoux A, B’chir W, Maurin AC, Müller S, Seibold F, Barnich N, Bruhat A, Darfeuille-Michaud A, Nguyen HT. Activation of the EIF2AK4-EIF2A/eIF2α-ATF4 pathway triggers autophagy response to Crohn disease-associated adherent-invasive Escherichia coli infection. Autophagy. 2016, 12(5):770-83. PMID: 26986695.
9. Nguyen HT, Dalmasso G, Mueller S, Carriere J, Seibold F, Darfeuille-Michaud A. Crohn’s disease-associated adherent-invasive Escherichia coli affect levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology. 2014, 146(2):508-19. PMID: 24148619.
10. Nguyen HT, Lapaquette P, Bringer MA, Darfeuille-Michaud A. Autophagy and Crohn’s disease. Journal of Innate Immunity. 2013, 5(5):434-43. PMID: 23328432.
11. Lapaquette P, Bringer MA, Darfeuille-Michaud A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cellular Microbiology. 2012, 14(6):791-807. PMID: 22309232.
12. Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hébuterne X, Harel-Bellan A, Mograbi B, Darfeuille-Michaud A, Hofman P. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s diseas. Nature Genetics. 2011, 43(3):242-5. PMID: 21278745.
13. Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cellular Microbiology. 2010; 12(1):99-113. PMID: 19747213.
Composition de l'équipe
Hang NGUYEN, CR Inserm, hang.nguyen@uca.fr
Emilie VAZEILLE, Ingénieur biologiste médical, emilie.vazeille@uca.fr
Guillaume DALMASSO, MCU, guillaume.dalmasso@uca.fr
Nicolas BARNICH (responsable de l'équipe M2iSH), Professeur des universités, nicolas.barnich@uca.fr
Anthony BUISSON, MCU-PH, a_buisson@chu-clermontferrand.fr
Laurène SALESSE, étudiante en thèse, laurene.salesse@uca.fr
Clara DOUADI, étudiante en thèse, clara.duadi@uca.fr
Alison DA SILVA, étudiante en thèse, alison.da_silva@uca.fr
Hanh HOANG, postdoctorante, thi_my_hanh.hoang@uca.fr