Cellular Microbiology and Physics of Infection
Center of Infection and Immunity of LilleCNRS UMR 8204 – INSERM U1019
Institut Pasteur de Lille – CHRU de Lille – Université de Lille
1, rue du Professeur Calmette, F-59019 Lille Cedex
- Lille
Site web - frank.lafont@pasteur-lille.fr -
Principal investigator
Frank Lafont

Research themes
According to the WHO 2005 world report, infectious diseases constitute the second leading cause for human mortality. New emerging pathogens and threat of outbreaks responsible of epidemic and pandemic leading to high rate of morbidity and mortality reinforce the research effort devoted to infectious agents. Also, the possibility of using pathogens for bioterrorism actions implies the development of new efficient therapeutic strategies.
We are interested in understanding the early steps of infection at the molecular and cellular levels. Our research focuses on the mechanisms hijacked by pathogens to interact and possibly invade the host cells. Pathogens have evolved many strategies to subvert molecular machineries of the host to escape immune response and manage a successful invasion.
We want to develop an interdisciplinary approach combining bacterial genetics, microbiology, cell and molecular biology, biochemistry and biophysics to investigate how pathogens interact with the host cell surface, how the cell responds to this attack and how pathogens subvert this response. The aim is to identify new therapeutic targets and to design new tools in order to inhibit infection.
The main topics are:
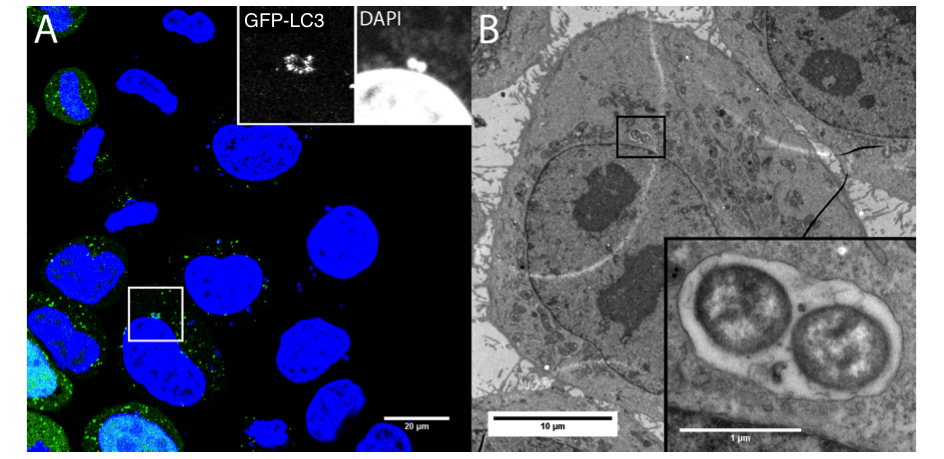
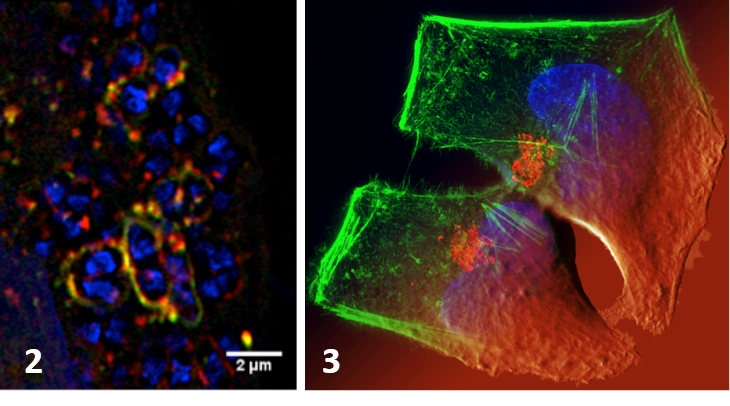
-The role of autophagy in the infectious mechanisms related to bacteria invasion of host cells. We study the control mechanisms of the bacterial vacuole involving its rupture and the membrane traffic modeling its regulation. We have shown the importance of autophagy in the cell response (incl. survival/death fate of the infected cell) to membrane damage linked to the vacuolar rupture after Shigella infection with the involvement of Galectins, adaptors and ubiquitination (Dupont et al. CHM 2009, Paz et al. Cellular Microbiol 2010). We have also shown ho Yersinia pseudotuberculosis hijacks the autophagy pathway to replicate in infected cells (Moreau et al. Cellular Microbiol 2010, Ligeon et al. Autophagy 2014). We are developing approaches using super-resolution imaging methods and correlatives microscopy (Ligeon et al. Methods 2015) to discriminate between signaling associated with interaction to the cell surface (adhesion) and entry (internalization).
-The role of biophysical properties of membranes in the control of the signal transduction during host-pathogen interactions. We analyze how interaction and adhesion forces, membrane or cellular elasticity, and viscosity participate to the regulation triggered by the interaction of a pathogen or bacterial toxins to the cell membranes (Mostowy et al Biophy J 2011 ; Maddugoda et al. CHM 2011 ; Gonzalez-Rodriguez et al. PRL 2012, Bernard et al. Nat Med 2014). We develop our approaches beyond the infectious theme as these cellular biomechanical aspects have implications on many different fields.
Descriptive figure
Publications
– Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, van der Goot FG, Sansonetti PJ, Lafont F. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. (2009) Cell Host Microbe. 6:137-49.
-Dupont N, Lafont F. How autophagy regulates the host cell signaling associated with the postpartum bacteria cocoon experienced as a danger signal. (2009) Autophagy. Nov;5(8):1222-3.
-Lafont F. Autophagy employs a new DAGger against bacteria. (2010) Cell Host & Microbe 8:129-130.
-Dupont N, Temime-Smaali N, Lafont F. How ubiquitination and autophagy participate in the regulation of the cell response to bacterial infection (2010) Biology of the Cell 102:621-634
-Paz I, Sachse M, Dupont N, Cederfur C, Enninga J, Leffler H, Poirier F, Prevost MC, Lafont F, Sansonetti P. Galectin-3, a marker for vacuole lysis by invasive pathogens. (2010) Cell Microbiol. 12 :530-544
-Moreau K, Lacas-Gervais S, Fujita N, Sebbane F, Yoshimori T, Simonet M, Lafont F. Autophagosomes can support Yersinia pseudotuberculosis replication in macrophages. (2010) Cell Microbiol. 12 :1108-1123.
-Mostowy S, Janel S, Forestier C, Roduit C, Kasas S, Pizarro-Cerda J, Cossart P, Lafont F A Role for Septins in the Interaction between the Listeria monocytogenes Invasion Protein InlB and the Met Receptor. (2011) Biophys. J. 100 :1949-1959
-Maddugoda MP, Stefani C, Gonzalez-Rodriguez D, Saarikangas J, Torrino S, Janel S, Munro P, Doye A, Prodon F, Aurrand-Lions M, Goossens PL, Lafont F, Bassereau P, Lappalainen P, Brochard F, Lemichez E. Anthrax co‐opts cAMP signaling to induce transendothelial cell tunnels, a Dewetting phenomenon counteracted by MIM (2011) Cell Host & Microbe. 10 : 464-474
-Nacer A, Roux E, Pomel S, Scheidig-Benatar C, Sakamoto H, Lafont F, Scherf A, Mattei D. Surface Exposure of Non-functional PfEMP1 in Plasmodium falciparum with Chromosome 9 Deletion is Not Linked to clag9 Expression (2011) PLoS ONE 6(12) :e29039
-Ligeon LA, Temime-Smaali N, Lafont F.Ubiquitylation and autophagy in the control of bacterial infections and related inflammatory responses (2011) Cellular Microbiol. 13:1303-1311.
-Klionsky D.J., (…) Lafont F et al. Guidelines for the use and interpretation of assays for monitoring autophagy (2012) Autophagy 8: 445-554.
-Gonzalez-Rodriguez D, Maddugoda MP, Stefani C, Janel S, Lafont F, Cuvelier D, Lemichez E, Brochard-Wyart F. Cellular Dewetting: Opening of Macroapertures in Endothelial Cells. (2012) Phys. Rev Letters , 208 :218105(5).
-Ligeon LA, Moreau K, Barois N, Bongiovanni A, Lacorre DA, Werkmeister E, Proux-Gillardeaux V, Galli T, Lafont F. Role of VAMP3 and VAMP7 in the commitment of Yersinia pseudotuberculosis to LC3-associated pathways involving single- or double-membrane vacuoles. (2014) Autophagy 10:1588-1602.
-Faure M, Lafont F. Pathogen-Induced Autophagy Signaling in Innate Immunity (2013) J Innate Immunity 5:456-470.
-Bernard SC, Simpson N, Join-Lambert O, Federici C, Laran-Chich MP, Maïssa N, Bouzinba-Ségard H, Morand PC, Chretien F, Taouji S, Chevet E, Janel S, Lafont F, Coureuil M, Segura A, Niedergang F, Marullo S, Couraud PO, Nassif X, Bourdoulous S. Pathgenic Neisserai meningitidis utilizes CD147 for vascular colonization. (2014) Nature Medicine 20:728-731
-Nollet M, Santucci-Darmanin S, Breuil V, Al-Sahlanee R, Cros C, Topi M, Momier D, Samson M, Pagnotta S, Cailleteau L, Battaglia S, Farlay D, Dacquin R, Barois N, Jurdic P, Boivin G, Heymann D, Lafont F, Lu SS, Dempster DW, Carle GF, Pierrefite-Carle V. Autophagy in Osteoblasts is involved in Mineralization and Bone Homeostasis (2014) Autophagy 10:1965-77
-Ligeon LA, Barois N, Werkmeister E, Bongiovanni A, Lafont F. Structured illumination microscopy and correlative microscopy to study autophagy (2015) Methods 75:61-68.
-Dubois-Deruy E, Belliard A, Mulder P, Bouveet M, Smet-Nocca C, Janel S, Lafont F, Beseme O, Amouyel P, Richard V, Pinet F. Interplay between troponin T phosphorylation and O-N-acetylglucosaminylation in ischaemic heart failure (2015) Cardiovasc. Res. 107:56-65
-Rodriguez-Emmenergger C., Janel S, de Los Santa Pereira A, Bruns M, Lafont F (2015) Quantifying bacterial adhesion on antifouling polymer brushes via single-cell force spectroscopy. Polymer Chem. 6, 5740-5751
Composition de l'équipe
Bauderlique Hélène, Ingénieure
Barois Nicolas, Ingénieur
Bongiovanni Antonino, Technicien
Bovio Simone, Post-doc
Ciczora Yann, Ingénieur
Dujardin Antoine, Thésard
Dupres Vincent, MCU Univ Lille 2
Janel Sébastien, Ingénieur
Popoff Michka, Thésard
Renaud Marie-Christine, Gestionnaire
Warein Joëlle, Technicienne
Werkmeister, Ingénieure