Genes-Nutrients (GN)
Gènes-Nutriments (GN)Université Clermont-Auvergne
Clermont-Ferrand - Clermont-Ferrand
Site web - pierre.fafournoux@inra.fr -
Principal investigator
Pierre Fafournoux

Research themes
The projects developed by the “Genes-Nutrients” team started from two observations: 1- Mammals are not able to synthesize essential amino acids (AA). Consequently, in case of AA scarcity, adaptation of the cellular and physiological functions is required. 2- The consequences of simple nutritional situations suggest that AAs play the role of “signal molecules” in these adaptive processes. These observations led us to ask the following question: Does amino acids regulate physiological functions through the regulation of gene expression?
At the creation of the team, the mechanisms by which AA-limitation led to the overexpression of a set of genes were not known. We contributed to the identification of a signaling pathway regulated by a lack of essential amino acid: the GCN2-eIF2α-ATF4 pathway.
In recent years the main projects developed in the team are the following:
– Identify the molecular mechanisms involved in the regulation of signaling pathways and gene expression in response to amino acid availability. In particular, we showed interactions between the pathways regulated by the kinases GCN2 and mTORC1 (Articles 2.6).
– Identify the cellular and physiological functions regulated by the GCN2-eIF2α-ATF4 pathway (autophagy and food intake) (Articles 5, 7, 8).
– Identify the role of eIF2α-ATF4 pathway in AA-deficient tumor cells (Article 1).
– Develop a nutritionally inducible gene regulation system for controlling transgene expression in gene therapy (Articles 3, 4).
– Study the consequences of protein malnutrition during the perinatal period of the life (Article 9).
Our research in nutrition has led us to study the regulation of autophagy. Indeed, in the case of dietary AA deficiency, a stimulation of proteolysis compensates the lack of AA and allows the maintenance of protein synthesis in the most sensitive organs. Literature describes a preponderant role of the inhibition of the protein kinase mTORC1 in the control of autophagy. However, the GCN2-eIF2α-ATF4 pathway being controlled by AA availability, we hypothesized that it could also be involved in the regulation of autophagy.
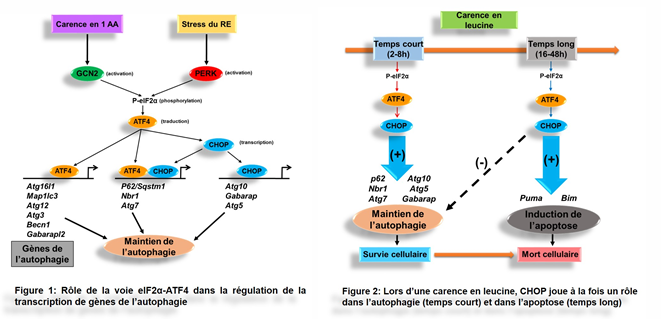
In a first step, we demonstrated that the eIF2α-ATF4 signaling plays a key role in regulating the expression of numerous genes involved in the autophagic process (Article 8). In particular, we identified three classes of autophagy genes according to their dependence on ATF4 and CHOP transcription factors and the binding of these factors on the specific elements of their promoters (Figure 1).
We have also shown that during the first six hours of the amino acid deficiency, the eIF2α-ATF4-CHOP pathway promotes cell survival by maintaining autophagy (Article 5). However, when the amino acid deficiency is prolonged (16-48h), CHOP plays a key role in the regulation of apoptosis and in the repression of the autophagic process by controlling the transcription of specific target genes (Figure 2).
In addition, we observed that leucine deficiency resulted in rapid activation of the autophagic process (<1h) in a GCN2-dependent manner. We therefore seek to determine the role of GCN2 in the regulation of early events of autophagy activation.
Descriptive figure
Publications
1- Mesclon F, Lambert-Langlais S, Carraro V, Parry L, Hainault I, Jousse C, Maurin AC, Bruhat A, Fafournoux P, Averous J. Decreased ATF4 expression as a mechaanism of acquired resistance to long-term amino acid limitation in cancer cells Oncotarget. 2017 in press.
2- Averous J, Lambert-Langlais S, Mesclon F, Carraro V, Parry L, Jousse C, Bruhat A, Maurin AC, Pierre P, Proud CG, Fafournoux P. GCN2 contributes to mTORC1 inhibition by leucine deprivation through an ATF4 independent mechanism. Sci Rep. 2016 Jun 14;6:27698.
3- Chaveroux C, Bruhat A, Carraro V, Jousse C, Averous J, Maurin AC, Parry L, Mesclon F, Muranishi Y, Cordelier P, Meulle A, Baril P, Do Thi A, Ravassard P, Mallet J, Fafournoux P. Regulating the expression of therapeutic transgenes by controlled intake of dietary essential amino acids. Nat Biotechnol. 2016 Jul;34(7):746-51.
4- Chaveroux C, Carraro V, Canaple L, Averous J, Maurin AC, Jousse C, Muranishi Y, Parry L, Mesclon F, Gatti E, Mallet J, Ravassard P, Pierre P, Fafournoux P, Bruhat A. In vivo imaging of the spatiotemporal activity of the eIF2α-ATF4 signaling pathway : insight into stress and related disorders. Sci Signal. 2015 Apr 28;8(374):rs5.
5- B’chir W, Chaveroux C, Carraro V, Averous J, Maurin AC, Jousse C, Muranishi Y, Parry L, Fafournoux P, Bruhat A. Dual role for CHOP in the crosstalk between autophagy and apoptosis to determine cell fate in response to amino acid deprivation. Cell Signal. 2014 Jul;26(7):1385-91.
6- Averous J, Lambert-Langlais S, Carraro V, Gourbeyre O, Parry L, B’Chir W, Muranishi Y, Jousse C, Bruhat A, Maurin AC, Proud CG, Fafournoux P. Requirement for lysosomal localization of mTOR for its activation differs between leucine and other amino acids. Cell Signal. 2014 Sep;26(9):1918-27.
7- Maurin AC, Benani A, Lorsignol A, Brenachot X, Parry L, Carraro V, Guissard C, Averous J, Jousse C, Bruhat A, Chaveroux C, B’chir W, Muranishi Y, Ron D, Pénicaud L, Fafournoux P. Hypothalamic eIF2α signaling regulates food intake. Cell Rep. 2014 Feb 13;6(3):438-44.
8- B’chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013 Sep;41(16):7683-99.
9- Jousse C, Parry L, Lambert-Langlais S, Maurin AC, Averous J, Bruhat A, Carraro V, Tost J, Letteron P, Chen P, Jockers R, Launay JM, Mallet J, Fafournoux P. Perinatal undernutrition affects the methylation and expression of the leptin gene in adults: implication for the understanding of metabolic syndrome. FASEB J. 2011 Sep;25(9):3271-8.