Dynamics of Cell Compartmentation in Plant
Institut des Sciences du Végétal , UPR 2355, CNRS.Imagif- Centre de Recherche de Gif sur Yvette, FRC 3115, CNRS.
(I2BC – Institut de Biologie Intégrative de la Cellule au 01/01/2015) - Gif sur Yvette
Site web - bsj@isv.cnrs-gif.fr -
Principal investigator
Béatrice Satiat-Jeunemaitre

Research themes
The group has a long standing experience in cell imaging of secretory trafficking pathways and in endomembrane biology in plant cells. It has recently outlined the interest of the plant cell as a model to decode autophagy mechanisms. On the other hand, photosynthetic organisms are also awaiting an answer for important questions in autophagy research.
The nature of the autophagosome is intriguing : a double membrane compartment, surrounding a portion of cytoplasm. As such, autophagosomes differ from the compartments of the endomembrane system, delimated by one membrane. This questions the formation mode of the autophagosome and its interactions with other membranous compartments. Indeed, while autophagosome formation is known to be associated to a complex cascade of signaling and molecular events involving ATG proteins, the precise positioning of these events in cellulo is not fully understood yet. The team aims to understand the mechanisms of autophagosome formation in plant cells and its role in plant adaptation to environmental stresses. Live Imaging of ATG proteins and multimodal microscopy approaches provide insight in the processes governing the morphogenesis of auphagosomal membranes, challenging or completing working hypotheses issued from other organisms. The group expertise in multiscale imaging analyses of the autophagy machinery allows collaborations with laboratories using other model systems (Djeddi et al., 2012; Manil-Segalen et al., 2014).
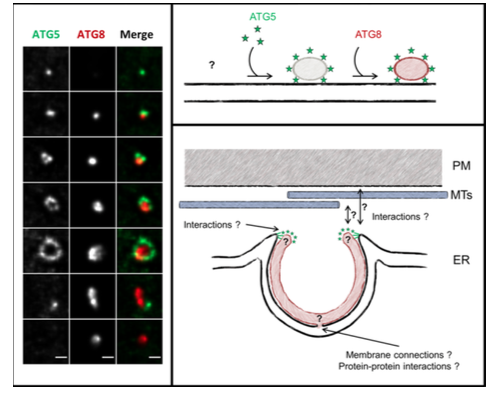
We have recently demonstrated in controlled starvation-induced conditions, a tight interaction of the forming autophagosome (the phagophore) with the underlying ER membranes (LeBars et al., 2014a, b).
The proposed model is reinforcing the apparently universal concept of ER membranes as template or feeder for the phagophore. We have shown that the ATG5 protein decorates the ER only in close proximity of the plasma membrane (PM), raising the question whether ER-PM contact sites could be dedicated platforms and/or membrane sources for the growing phagophore. Moreover, additional questions wait for answers: what are the mechanisms governing this ER/phagophore early association, and close contact with the ER? How does the phagophore grow into a cup-like structure? Is there a specific “ER autophagosome formation site (“ERAFS”) marked by specific lipid and protein signature? We tackle these questions in an integrative approach of autophagosome morphogenesis in plant cells, using the lab expertise in cell biology , cell imaging, and plant cell biology, and will be more than happy to mutualise this effort in autophagy research with other “autophagy teams”.
Descriptive figure
Publications
– Le Bars, R., Marion, J., Le Borgne, R., Satiat-Jeunemaitre, B. & Bianchi, MW. (2014) ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 5, 4121 (2014).
– Djeddi, A., Michelet, X., Culetto, E., Alberti, A., Barois, N. & Legouis, R. (2012) Induction of autophagy in ESCRT mutants is an adaptive response for cell survival in C. elegans. Journal of cell science 125, 685-694.
– Le Bars R., Marion J., Satiat-Jeunemaitre B., Bianchi MW. (2014). Folding into an autophagosome : Atg5 sheds
light on how plants do it. Autophagy 10, 1-3
– Manil-Ségalen M, Lefebvre C, Jenzer C, Trichet M, Boulogne C, Satiat-Jeunemaitre B, Legouis R. (2014) The C.
elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39.Dev Cell. 28(1):43-55 Erratum in: Dev Cell. 2014 Jul 14;30(1):110.
Composition de l'équipe
Groupe « Morphodynamique du système endomembranaire »
Michele Wolfe Bianchi (MCU UPEC) (1)
Romain le Bars (PhD) (2)
Jessica Marion (AI, CNRS) (3)
Frederic Coquelle (MCU Paris-Sud) (4)
Arthur Molines (Etudiant en thèse)
Mebarek Temagoult (AI, CDD) (5)
Groupe “Pôle Imagerie et Biologie Cellulaire d’Imagif”
Claire Boulogne (IR) (6)
Mickael Bourge (IE) (7)
Cynthia Gillet (AI) (8)
Romain LeBars (CDD, IE) (9)
Laetitia Besse (CDD, IE) (10)