Contrôle de la croissance par les nutriments
Inserm U1151, Institut Necker Enfants MaladesFaculté de Médecine Paris Descartes, Bat. Leriche, porte 9
14 Rue Maria Helena Vieira Da Silva, CS61431
75993 Paris cedex 14 - Paris
Site web - mario.pende@inserm.fr - 0172606386
Principal investigator
Mario Pende

Research themes
In metazoans, nutrient and growth factor availability control cell number, size and metabolic homeostasis. We investigate the specific programs underlying these responses, and their coordination by signal transduction mechanisms.
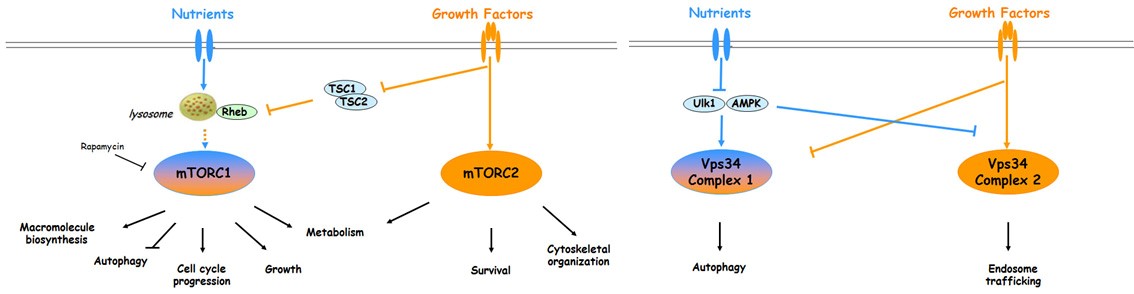
We focus on two nutrient signal transduction pathways, the mTOR (mammalian Target Of Rapamycin) and the Vps15/Vps34 complex (Vacuolar Protein Sorting15/34). These ancient pathways are present in every eukaryotic cell, from unicellular organisms like yeast to humans. They function as essential mechanisms that direct how growth and metabolism adapt to nutritional cues. mTOR is a Ser/Thr protein kinase, while the Vps15/Vps34 complex is a lipid kinase with phosphatidylinositol 3-kinase activity (class III PI3K). The transduction mechanisms triggered by mTOR and class III PI3K are complex. In mammalian cells, both kinases are engaged in multiple complexes that have different localization, targets and sensitivities to upstream signals, like nutrients and insulin.
We have contributed to demonstrate that this crosstalk and differential regulation may explain many physiological responses to nutrition. For instance, why nutrients and insulin are synergistic for cell growth, though nutrients cause resistance to the metabolic action of insulin. Or why insulin inhibits autophagy, though stimulates receptor trafficking.
During the past fifteen years we have generated and characterized a wide panel of mouse mutants in the mTOR and class III PI3K pathways. We were involved in revealing unique and interesting phenotypes that increased our knowledge of mTOR/class III PI3K roles in pathophysiology: mutants with small cells (Pende et al., Nature, 2000; Ohanna et al., Nature Cell Biol, 2005), mutants with impaired ribosome biogenesis (Fumagalli et al., Nature Cell Biol, 2009), mutants resistant to tumorigenesis in specific tissues and after specific oncogenic insults (Alliouachene et al., JCI, 2008; Panasyuk et al., Nature Comm., 2012; Patitucci et al., JCI, 2017), mutants with muscle disease (Risson et al., JCB, 2009; Nemazanyy et al., EMBO Mol Med, 2013), mutants mimicking caloric restriction and promoting longevity (Aguilar et al., Cell Metabolism, 2007; Barilari et al., EMBO J, 2017), mutants with altered insulin action (Nemazanyy et al., Nature Comm., 2015; Treins et al., Mol Cel Biol, 2012).
Au cours des quinze dernières années, nous avons généré et caractérisé un large panel de souris mutantes dans les voies mTOR et PI3K de classe III. Nous avons été impliqués dans la révélation de phénotypes uniques et intéressants qui ont augmenté notre connaissance des rôles de mTOR / PI3K de classe III en physiopathologie: des mutants avec de petites cellules (Pende et al., Nature, 2000, Ohanna et al., Nature Cell Biol, 2005), des mutants de la biogenèse des ribosomes (Fumagalli et al., Nature Cell Biol, 2009), des mutants résistants à la tumorigenèse dans des tissus spécifiques et après des insultes oncogéniques spécifiques (Alliouachene et al., JCI, 2008, Nature Comm. Patitucci et al., JCI, 2017), des mutants avec une maladie autophago-lysosomale (Nemazanyy et al., EMBO Mol Med, 2013), des mutants mimant la restriction calorique et favorisant la longévité (Aguilar et al. Metabolism, 2007, Barilari et al., EMBO J, 2017), des mutants avec une action modifiée de l’insuline (Nemazanyy et al., Nature Comm., 2015, Treins et al., Mol Cel Biol, 2012).
Descriptive figure
Publications
– Patitucci C., Couchy G., Bagattin A., Cañeque T., de Reyniès A., Scoazec J-Y, Rodriguez R., Pontoglio M., Zucman-Rossi J., Pende M.* and Panasyuk G. (2017) *corresponding author. HNF1α suppresses steatosis associated liver cancer by inhibiting PPARγ transcription. Journal of Clinical Investigation, 127, 1873-1888, doi: 10.1172/JCI90327.
– Barilari M., Bonfils G., Treins C., Koka V., De Villeneuve D., Fagrega S., Pende M. (2017) ZRF1 is a novel S6 kinase substrate that drives the senescence program. EMBO J., 36, 736-750. doi: 10.15252/embj.201694966.
– Nemazanyy I., Montagnac G., Russell R.C., Morzyglod L., Burnol A.F., Guan K.L., Pende M. *, Panasyuk G. (2015) *corresponding author .Class III PI3K regulates organismal glucose homeostasis by providing negative feedback on hepatic insulin signalling. Nature Communications, 21, 8283. doi: 10.1038/ncomms9283.
– Liang N., Zhang C., Dill P., Panasyuk G., Pion D., Koka V., Gallazzini M., Olson E.N., Lam H., Henske E.P., Dong Z., Apte U., Pallet N., Johnson R.L., Terzi F., Kwiatkowski D.J., Scoazec J-Y., Martignoni G., Pende M. (2014) Regulation of YAP by mTOR and autophagy reveals a therapeutic target of tuberous sclerosis complex. J Exp Med. 211, 2249-2263. doi: 10.1084/jem.20140341.
– Nemazanyy I., Blaauw B., Paolini C., Caillaud C., Protasi F., Mueller A., Proikas-Cezanne T., Russell R.C., Guan K-L., Nishino I., Sandri M., Pende M.*, Panasyuk G. (2013) *corresponding author. Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Mol Med, 5, 870-890. doi: 10.1002/emmm.201202057.
– Chauvin C., Koka V., Nouschi A., Mieulet V., Hoareau-Aveilla C., Dreazen A., Cagnard N., Carpentier W., Kiss T., Meyuhas O., Pende M. (2013) Ribosomal protein S6 kinase activity controls the Ribosome Biogenesis transcriptional program. Oncogene, 33, 474-483. doi: 10.1038/onc.2012.606. Epub 2013 Jan 14.
– Treins C., Alliouachene S., Hassouna R., Xie Y., Birnbaum M.J., Pende M. (2012) The combined deletion of S6K1 and Akt2 deteriorates glycemic control in a high-fat diet. Mol Cell Biol, 32, 4001-4011.
– Panasyuk G., Espeillac C., Chauvin C., Pradelli L.A., Horie Y., Suzuki A., Annicotte J.S., Fajas L., Foretz M., Verdeguer F., Pontoglio M., Ferré P., Scoazec J.Y., Birnbaum M.J., Ricci J.E., Pende M. (2012) PPARγ contributes to PKM2 and HK2 expression in fatty liver. Nature Communications, 3:672. doi: 10.1038/ncomms1667.
– Espeillac C., Mitchell C., Celton-Morizur S., Chauvin C., Koka V., Gillet C., Albrecht J.H., Desdouets C., Pende M. (2011) S6 kinase 1 activity is required for rapamycin-sensitive liver proliferation after mouse hepatectomy. Journal of Clinical Investigation, 121, 2821-2832.
– Alliouachene S., Tuttle R.L., Boumard S., Lapointe T., Berissi S., Germain S., Jaubert F., Tosh D., Birnbaum M.J., Pende M. (2008) Constitutively active Akt1 expression in mouse pancreas requires S6 Kinase 1 for insulinoma formation. Journal of Clinical Investigation, 118, 3629-3638.
– Aguilar V., Alliouachene S., Sotiropoulos A., Sobering A., Athea Y., Djouadi F., Miraux S., Thiaudière E., Foretz M., Viollet B., Diolez P., Bastin J., Benit P., Rustin P., Carling D., Sandri M., Ventura-Clapier R., Pende M. (2007) S6 kinase deletion suppresses muscle growth adaptations to nutrient availability by activating AMP kinase. Cell Metabolism, 5, 476-487.
– Ohanna M., Sobering A.K., Lapointe T., Lorenzo L., Praud C., Petroulakis E., Sonenberg N., Kelly P.A., Sotiropoulos A., Pende M. (2005) Atrophy of S6K1-/- skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat. Cell Biol. 7, 286-294.
Composition de l'équipe
Ganna Panasyuk, CR1 Inserm
Anne Sophie Armand, MCU Paris Descartes University
Catherine Caillaud, MCU-PH, Hopital Necker
Muriel Girard, MCU-PH, Hopital Necker
Vonda Koka, Ingenieur IE, Inserm
Delphine de Villeneuve, Ingenieur AI, Paris Descartes University
Martina Bonucci, PhD student
Nicolas Kuperwasser, Postdoc
Talha Rashid, PhD student
Ning Liang, Postdoc
Anton Iershov, Postdoc
Chantal Alkhoury, PhD student,
Paul Crespin, M2 Student
Tiffen El Belaidi, M2 Student
Camille Streiff, M2 Student
Edward Chaloui, M2 Student