Wine-Food-Microbiology and Stress (VALMIS)
UMR PAM, Esplanade Erasme, 21 000, DIJON - DijonSite web - pierre.lapaquette@u-bourgogne.fr -
Principal investigator
Pierre Lapaquette

Research themes
The VALMIS team is interested in studying microbes of industrial interest (yeast and bacteria used for food processing or used as probiotics in human and animal care) and especially their interactions with environment (host organism, other microbes, wine and food matrix).
A first part of the team is focused on molecular mechanisms used by microbes during wine making to face hostile conditions arising during this industrial process (including acidification, presence of ethanol, low nutrient availability). They also investigate relationships existing between wine-associated microbial communities and wine alterations linked to the development of deleterious microbes.
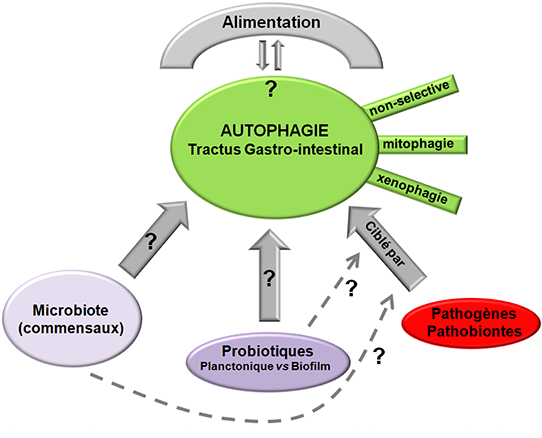
A second part of the team, including clinicians, is interested in bacterial and yeast probiotics. Probiotics are defined as live microorganisms that, when administered in adequate amounts, confer a health benefit on the host. Our team explores stress resistance mechanisms induced by probiotics microbes during their transit through the gastrointestinal tract and the ability of these microbes to secrete biologically active molecules interacting with the host immune system or with the intestinal microbiota. Effects of these probiotics are assessed in various disease states including inflammatory bowel diseases and infection (with gut pathogens such as Salmonella spp. or Candida albicans).
A recent research topic of our team is to develop probiotics microorganisms (natural or genetically engineered strains), or innovative formulation of these microbes, able to sustain intestinal homeostasis, especially by stimulating the autophagic process. To date, activation of autophagy by pathogenic microorganisms and their subsequent elimination by this process have been extensively studied. Far less is known regarding the regulation of autophagy by the whole intestinal microbiota, individual commensal microorganisms or probiotics. Thus, our research project focus on two main axis: (1) to understand the role played by the whole intestinal microbiota on autophagy during basal state and during disease states (infection or inflammatory bowel diseases) and (2) to select commensal, probiotic strains or to genetically engineer bacterial strains to enhance autophagic activity in the gut, and thereby reinforcing intestinal homeostasis. We characterize the effects of these microorganisms on the autophagic process in vitro by using polarized intestinal epithelial cells lines and in vivo using mice (germ free vs conventional) and zebra fish model organism.
Descriptive figure
Publications
– Aoudia N., Rieu A., Briandet R., Deschamps J., Chluba J., Jego G., Garrido C., Guzzo J. (2016). Biofilms of Lactobacillus plantarum and Lactobacillus fermentum: effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiology, 53, 51-59.
– Goyer M, Loiselet A, Bon F, L’Ollivier C, Laue M, Holland G, Bonnin A, Dalle F (2016). Intestinal Cell Tight Junctions Limit Invasion of Candida albicans through Active Penetration and Endocytosis in the Early Stages of the Interaction of the Fungus with the Intestinal Barrier. PLoS One. 2016 Mar 2;11(3):e0149159.
– Albac S, Schmitz A, Lopez-Alayon C, d’Enfert C, Sautour M, Ducreux A, Labruère-Chazal C, Laue M, Holland G, Bonnin A, Dalle F (2016). Candida albicans is able to use M cells as a portal of entry across the intestinal barrier in vitro. Cell Microbiol. 2016 Feb;18(2):195-210.
– Bastard A, Coelho C, Briandet R, Canette A, Gougeon R, Alexandre H, Guzzo J, Weidmann S (2016) Effect of Biofilm Formation by Oenococcus oeni on Malolactic Fermentation and the Release of Aromatic Compounds in Wine. Front Microbiol. 2016 Apr 27;7:613.
– Lapaquette P, Guzzo J, Bretillon L and Bringer MA. Cellular and molecular connections between autophagy and inflammation. Mediators Inflamm. 2015;2015:398483.
– Darsonval M, Msadek T, Alexandre H, Grandvalet C. (2015)The Antisense RNA Approach: a New Application for In Vivo Investigation of the Stress Response of Oenococcus oeni, a Wine-Associated Lactic Acid Bacterium. Appl Environ Microbiol. 2015 Oct 9;82(1):18-26.
– Grangeteau C, Daniel Gerhards, Sébastien Terrat, Samuel Dequiedt, Hervé Alexandre, Michèle Guilloux-Bénatier, Christian von Wallbrunn, Sandrine Rousseaux (2015) FT-IR spectroscopy: a powerful tool for studying the inter- and intraspecific biodiversity of cultivable yeasts isolated from grape must. Journal of Microbiological Methods 121, 50-58.
– Youzhong L, Rousseaux S, Tourdot-Maréchal R, Sadoudi M, Gougeon G, Schmitt-Kopplin P, Alexandre H (2015). Wine microbiome, a dynamic world of microbial interactions. Critical Reviews in Food Science and Nutrition. 2015 Jun 11:0.
– Rieu A, Aoudia N, Jego G, Chluba J, Yousfi N, Briandet R, Deschamps J, Gasquet B, Monedero V, Garrido C, Guzzo J (2014). The biofilm mode of life boosts the anti-inflammatory properties of Lactobacillus. Cell Microbiol. 2014 Dec;16(12):1836-53.
– Maitre M, Weidmann S, Dubois-Brissonnet F, David V, Covès J, Guzzo J (2014). Adaptation of the wine bacterium Oenococcus oeni to ethanol stress: role of the small heat shock protein Lo18 in membrane integrity. Appl Environ Microbiol. 2014 May;80(10):2973-80.
– Maitre M, Weidmann S, Rieu A, Fenel D, Schoehn G, Ebel C, Coves J, Guzzo J (2012). The oligomer plasticity of the small heat-shock protein Lo18 from Oenococcus oeni influences its role in both membrane stabilization and protein protection. Biochem J. 2012 May 15;444(1):97-104.
– Lapaquette P, Bringer MA, Darfeuille-Michaud A (2012). Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell Microbiol. 2012 Jun;14(6):791-807.
– Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hébuterne X, Harel-Bellan A, Mograbi B, Darfeuille-Michaud A, Hofman P. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011 Mar;43(3):242-5.
Composition de l'équipe
Jana Al Azzaz (Doctorante 2ème année)
Hervé Alexandre (PU, Directeur Valmis)
Louise Basmacyan (Ass. Hospitalo-Universitaire)
Fabienne Bon (MCU)
Antonio Camara (Doctorant 1er année)
Frédéric Dalle (PU-PH, Chef de service parasitologie/mycologie, CHU Dijon)
Vanessa David-Vaizant (AI)
Amandine Ducreux (Tech)
Antoine Gobert (Doctorant 1er année)
Cosette Grandvalet (MCU)
Jean Guzzo (PU)
Michèle Guilloux-Benatier (MCU)
Corrine Hatier (Secrétaire)
Arnaud Heumann (Doctorant 2ème année)
Pierre Lapaquette (MCU)
Julie Laurent (Tech)
Alicia Loiselet (Doctorante 2ème année)
Cédric Longin (Doctorant 3ème année)
Luc Perepelkine (Ingénieur d’étude)
Clément Petitgonnet (Doctorante 1ère année)
Aurélie Rieu-Guigon (MCU)
Sandrine Rousseaux (MCU)
Raphaëlle Tourdot-Maréchal (MCU)
Marc Sautour (MCU-PH)
Stéphanie Weidmann-Desroches (MCU)